A new role of CBs in pre-mRNA metabolism
The life course of mRNA begins with transcription by RNA Pol II, splicing, and processing 5′ and 3′ ends, which generally is co-transcriptional. It enables efficient and fast transport to the cytoplasm, where the information will be read in the translation process. Typically, the time that elapses from the pre-mRNA transcript synthesis to the protein’s formation in the cytoplasm is short and lasts from a few to tens of minutes. Until recently, it was thought that expression of a given gene is mainly influenced by the level of transcription, transcripts stability (half-life of mRNA in the cytoplasm) and time of occurrence in the cell of the final product – the protein. However, some studies have shown that in specific cell types, it is observed that a significant portion of polyadenylated transcripts are “nuclear retained” and undetected in the cytoplasm. Thus they are not translated immediately after synthesis. Because pre-mRNA undergoes modifications during the synthesis of RNA (co-transcriptionally), fully mature mRNA can be formed relatively quickly. Therefore, this long retention of nuclear mRNA suggests that the nucleus exhibits an additional role previously skipped. Studies on the nuclear retention of the mRNA indicate that it significantly impacts gene expression by regulating the export and translation delay, which allows the synthesis of specific proteins under strictly controlled conditions and time.
Regulation of gene expression by nuclear retention of the mRNA had been demonstrated, among others, in the mammalian metabolic tissues, in generative cells or the cells under stress conditions. Although the final result of such regulation is always the same, the transcripts can be retained in different forms. These transcripts may be retained as mature mRNA or pre-mRNA. Some mRNAs have additional sequences that regulate the transport from the nucleus to the cytoplasm. These sequences are present in the regions of untranslated mRNA – 5 ‘UTR and 3’ UTR. There is also a hypothesis that nuclear retention factors bind the mRNAs and block the possibility of binding the export factors. It is suggested that a significant effect on the retention mRNA can have splicing factors that form an early spliceosome complex. It is also expected that these factors anchor mRNA in some nuclear structures.
Much less is known about the spatial organization of the process in the cell nucleus. So far, the only described domains associated with the accumulation of polyadenylated transcripts are nuclear speckles. However, accumulated in these structures, transcripts do not seem to be exported to the cytoplasm. In larch microsporocytes, during diplotene, we observed first accumulation of poli(A)RNA (Kolowerzo et al. 2009) and than mRNA (Smolinski and Kolowerzo 2012) in the nucleus in the Cajal bodies. These nuclear domains “retain” polyadenylated transcripts for a very long time.
Two patterns of mRNA nuclear retention and export to cytoplasm were observed. The majority of the studied transcripts followed the first one, consisting of a long retention period and slow release to the cytoplasm. The transcript encoding subunit 10 of RNA pol II was regulated according to the first pattern, which might constitute a mechanism ensuring tight transcription control due to negative feedback. mRNAs encoding translation factors and SERRATE followed the second pattern, in which the retention period was shorter, and transcripts were rapidly transferred to the cytoplasm (Majewska et al. 2021).
CBs are evolutionarily conserved structures occurring in both animal and plant cells. This indicates their fundamental role in the nuclear metabolism of eukaryotic cells. Intensive research, carried out for many years, has demonstrated the relationship between CBs and many processes associated with the metabolism of various types of RNA, for example, the maturation and assembly of snRNP (Smolinski et al. 2011; Hyjek et al. 2015), recruitment and assembly in ribosomal RNA maturation, tRNA maturation, histone mRNA maturation, the synthesis of telomeres and miRNA and siRNA maturation.
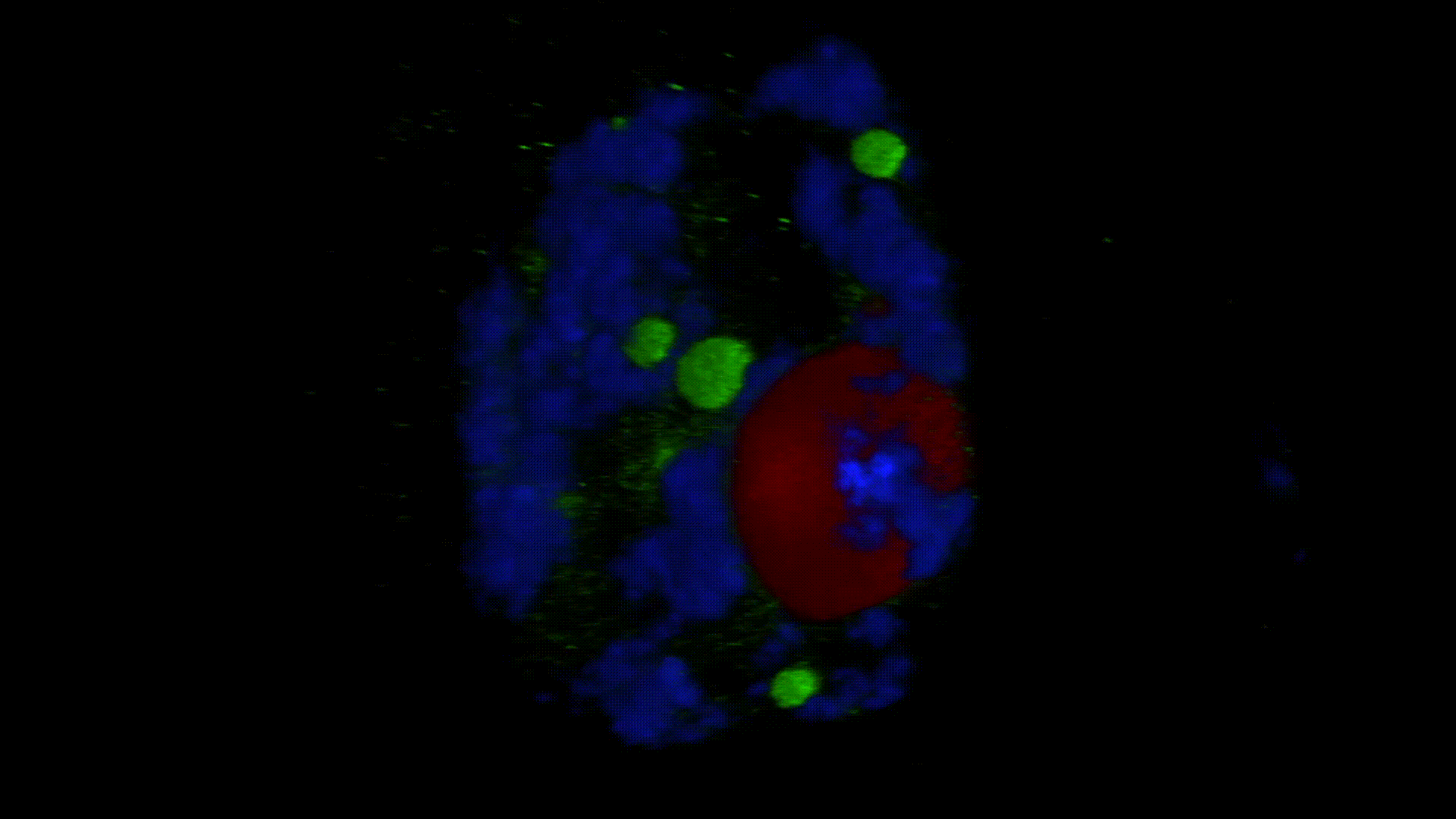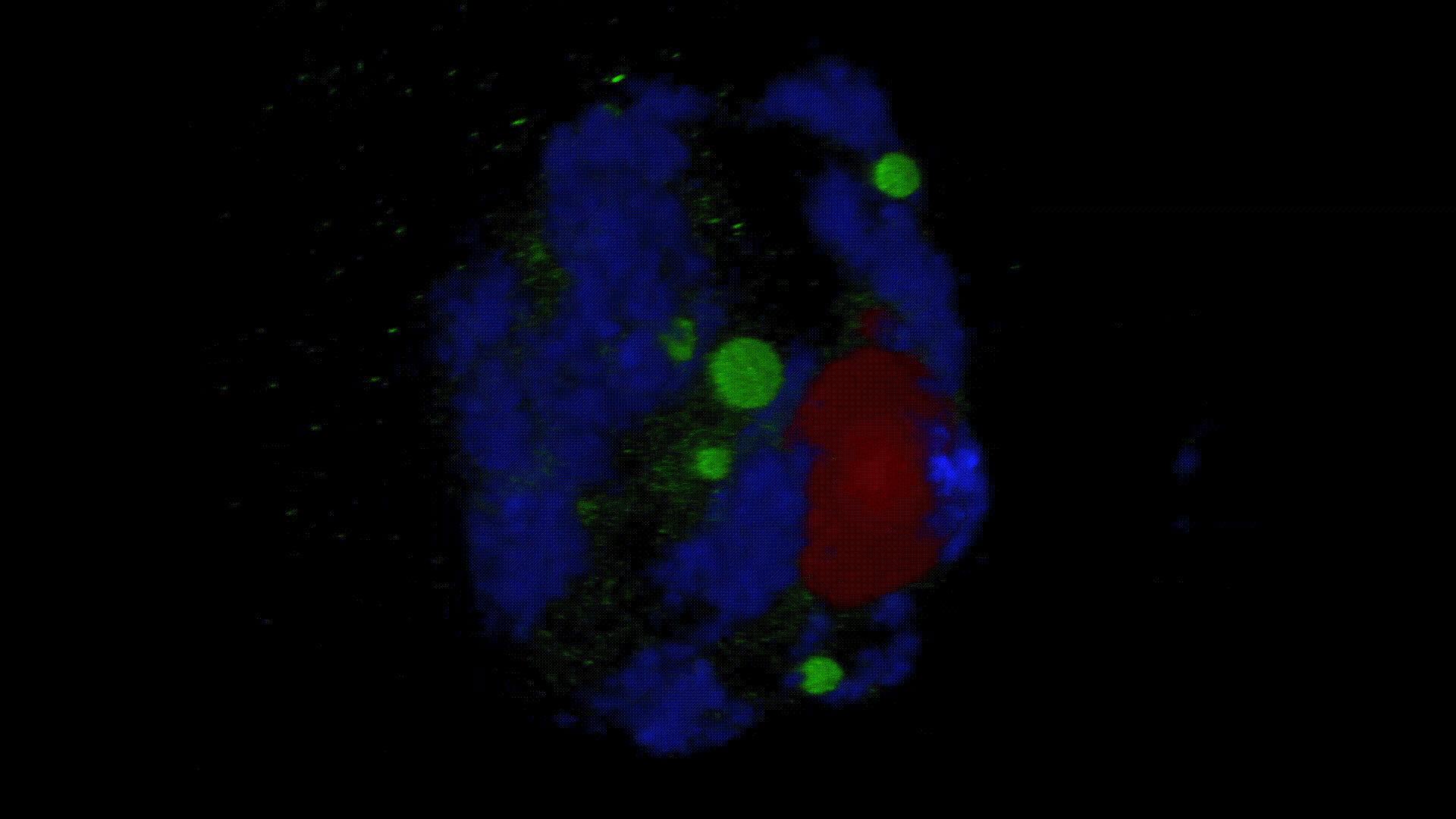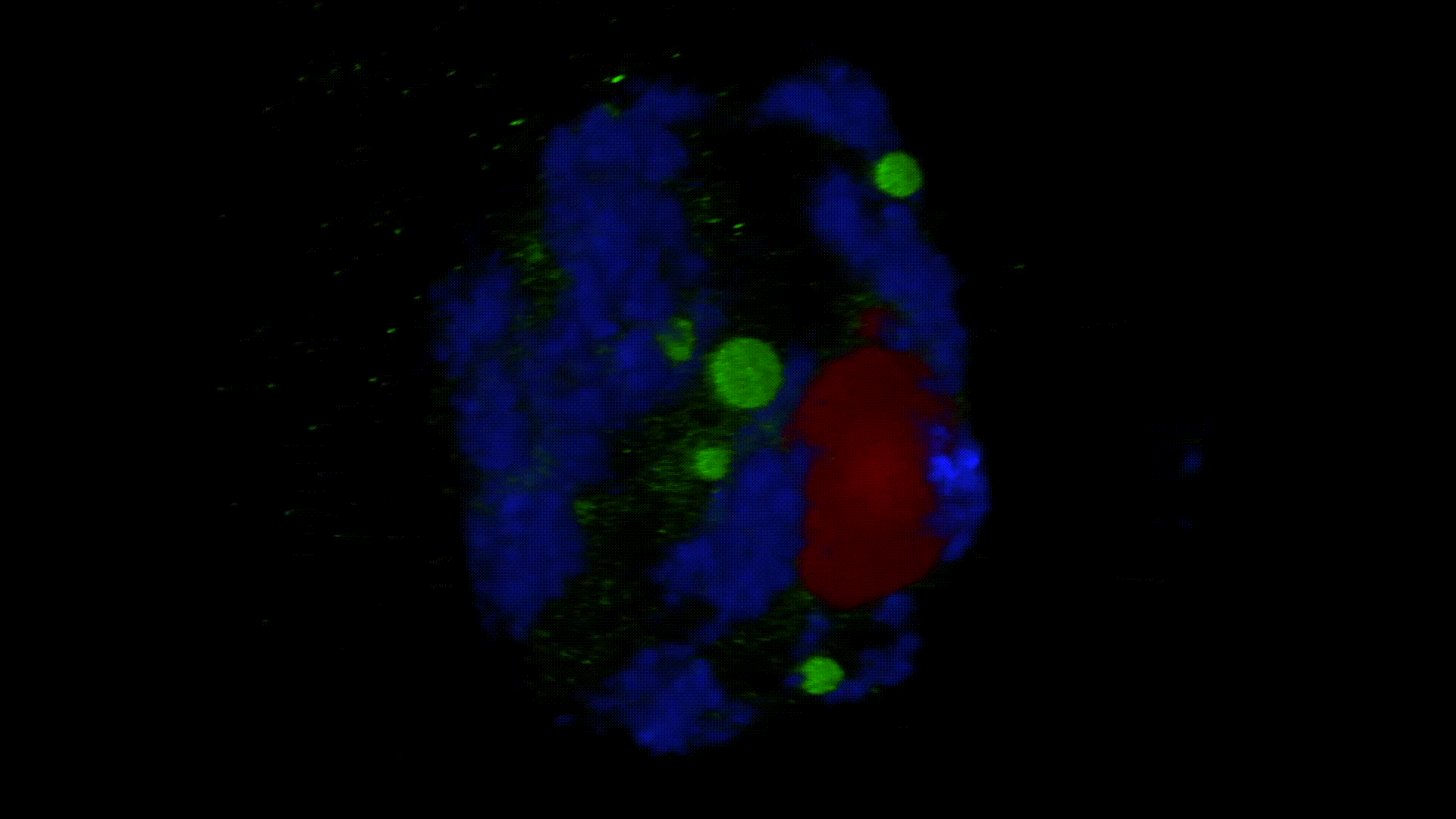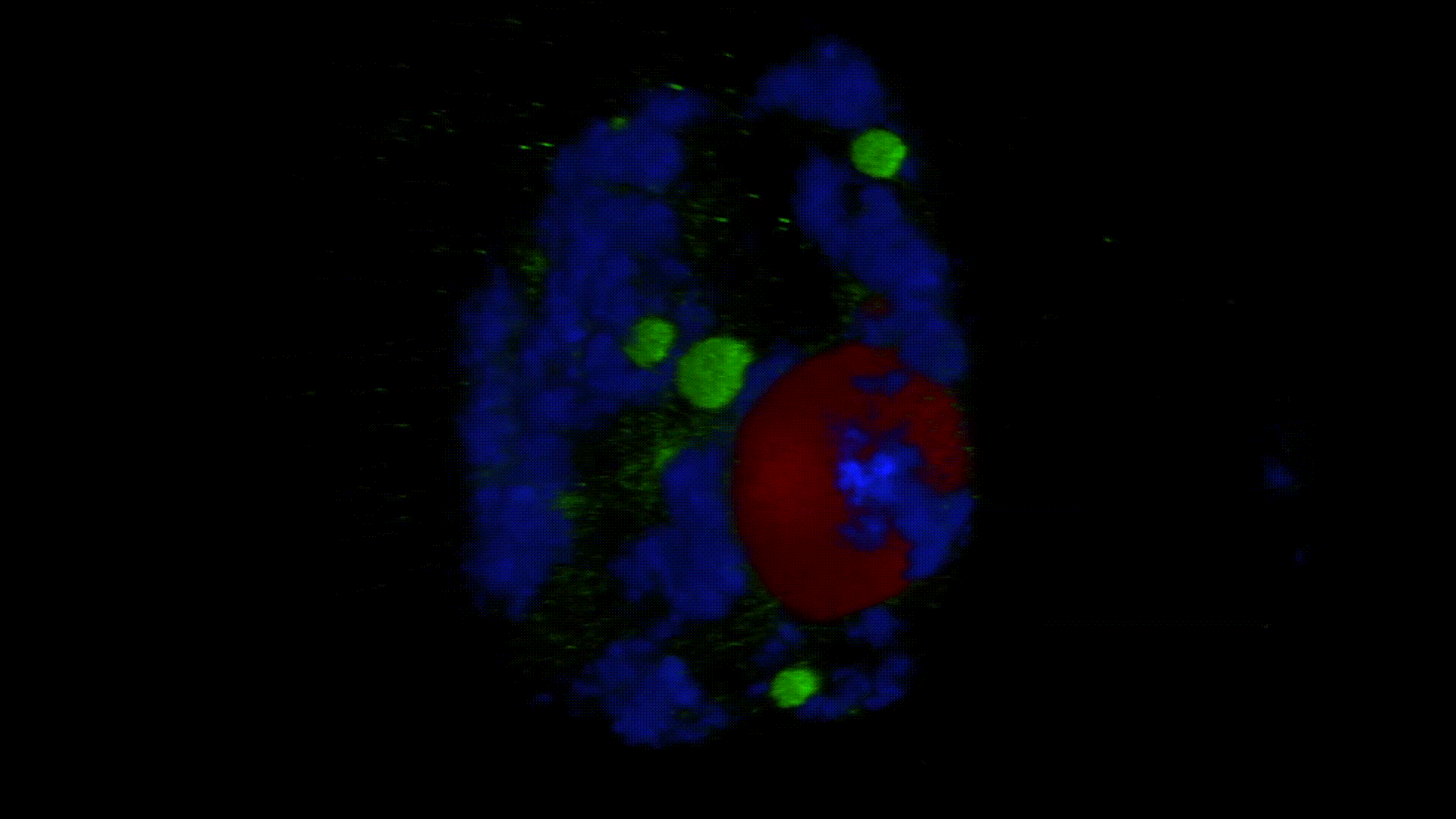
CBs are self-organized structures, appearing on local needs of increased metabolism of certain types of RNA. Assembling spliceosomal subunits is possible in the nucleoplasm, but it has been demonstrated that the process is carried out with more than 30-fold greater efficiency in the CB. For example fibroblast, cells characterized by low cellular metabolism do not possess CBs. Coilin gene knockdown (marker protein of those bodies) in cells with a high metabolism (generative cells or embryonic cells) causes the breakdown of CBs and leads to the death of these cells.
In order to verify that Cajal bodies are involved in post-transcriptional regulation of gene expression by nuclear retention of mRNAs or pre-mRNAs transcripts, we are currently conducting research on which mRNAs are stored in CBs, as well as we are trying to explain what mechanism is responsible for the retention of the mRNAs in CBs (Rudzka et al. 2021, Majewska et al. 2021, Rudzka et al. 2022).
To achieve these goals, we use laser microdissection (video4), analysis of the transcriptome of larch microsporocytes, proteomic analysis of proteins present in the cytoplasm of microsporocytes, bioimaging techniques in situ and in vivo at the level of single RNA molecules and PLA technique.
We believe that the retention may undergo both transcripts of genes involved in the meiotic division and the housekeeping gene transcripts. Understanding this phenomenon will significantly expand our better understanding of the processes of post-transcriptional regulation of gene expression, Cajal bodies role in the metabolism of RNA and mechanisms involved in the maturation of generative plant cells. It will enrich the current knowledge on cell biology as well as developmental biology.

 ul. Lwowska 1, 87-100 Toruń
ul. Lwowska 1, 87-100 Toruń